TASUKE
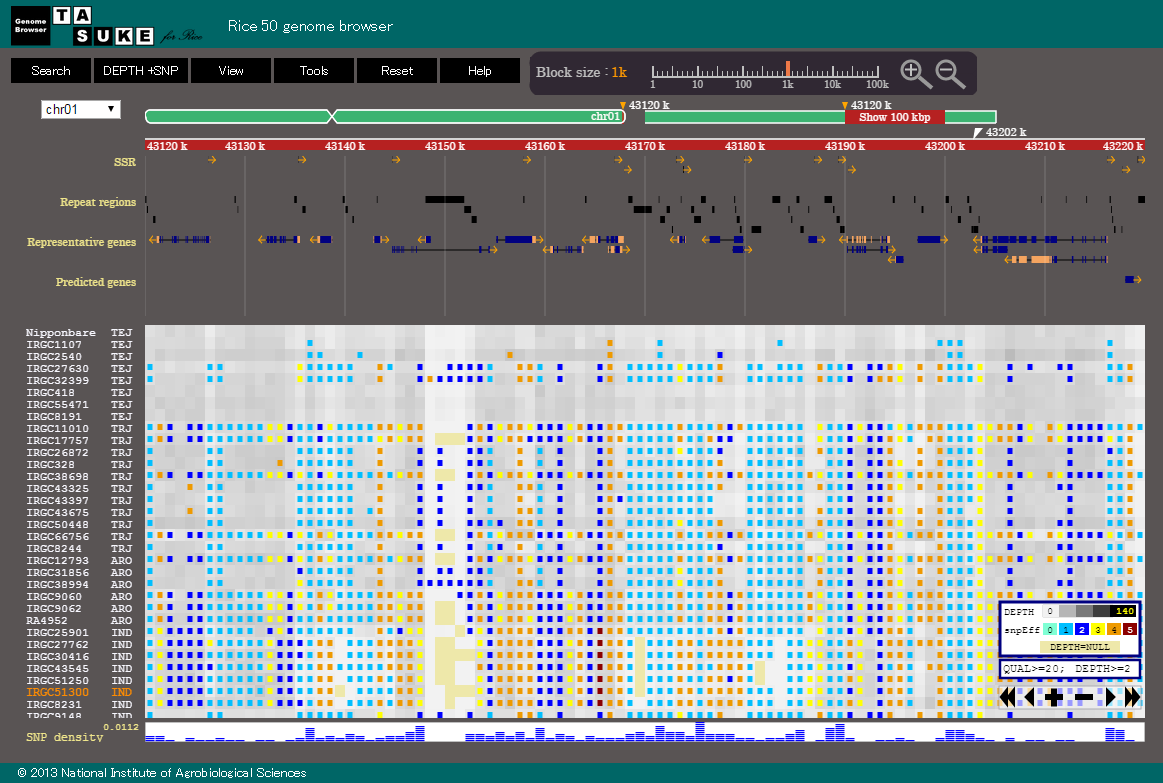
TASUKE is a web-based genome browser that visualizes large-scale resequencing data generated by next-generation sequencing technologies.
The variation and read depth of multiple genomes, as well as annotations, can be shown simultaneously at various scales.
The frequency of Variants and Depth of coverage are shown by gradational colors in a block whose size is variable from 1bp to 100kb.
Maximally 200 blocks can be shown at one window, so that up to 20 Mb can be viewed.
How to Get & InstallFollow these 4 steps and get TASUKE browser with your own data. |
|
Contact UsAsk any questions on installing, and we will support you. Bug report, suggestions and any comments are also welcome!! E-mail: |