Functions
Screen Shots
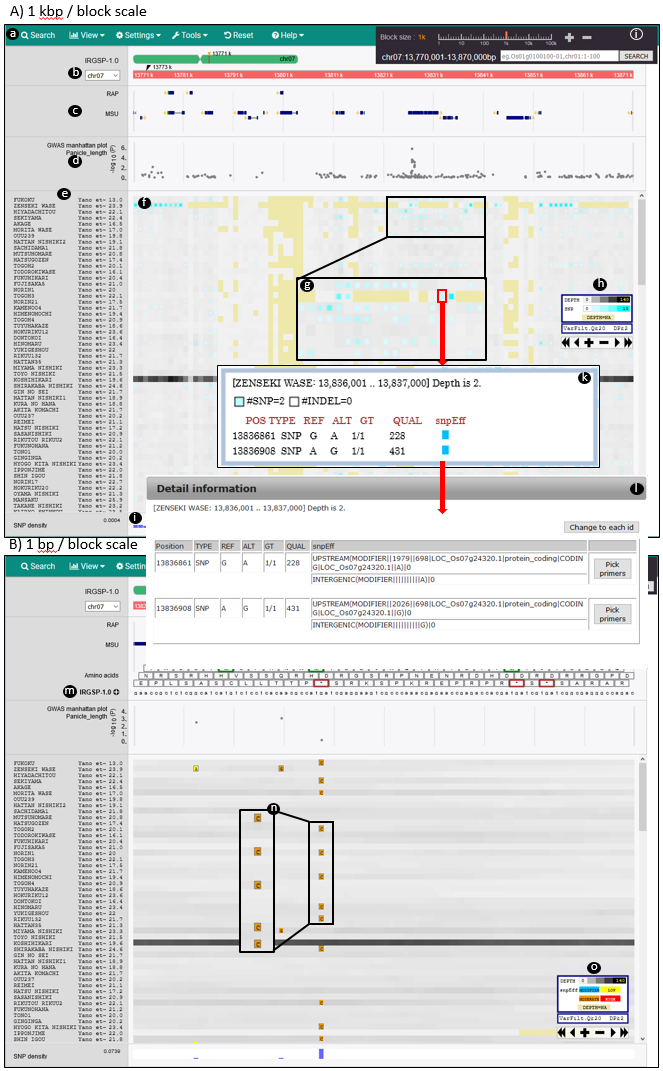
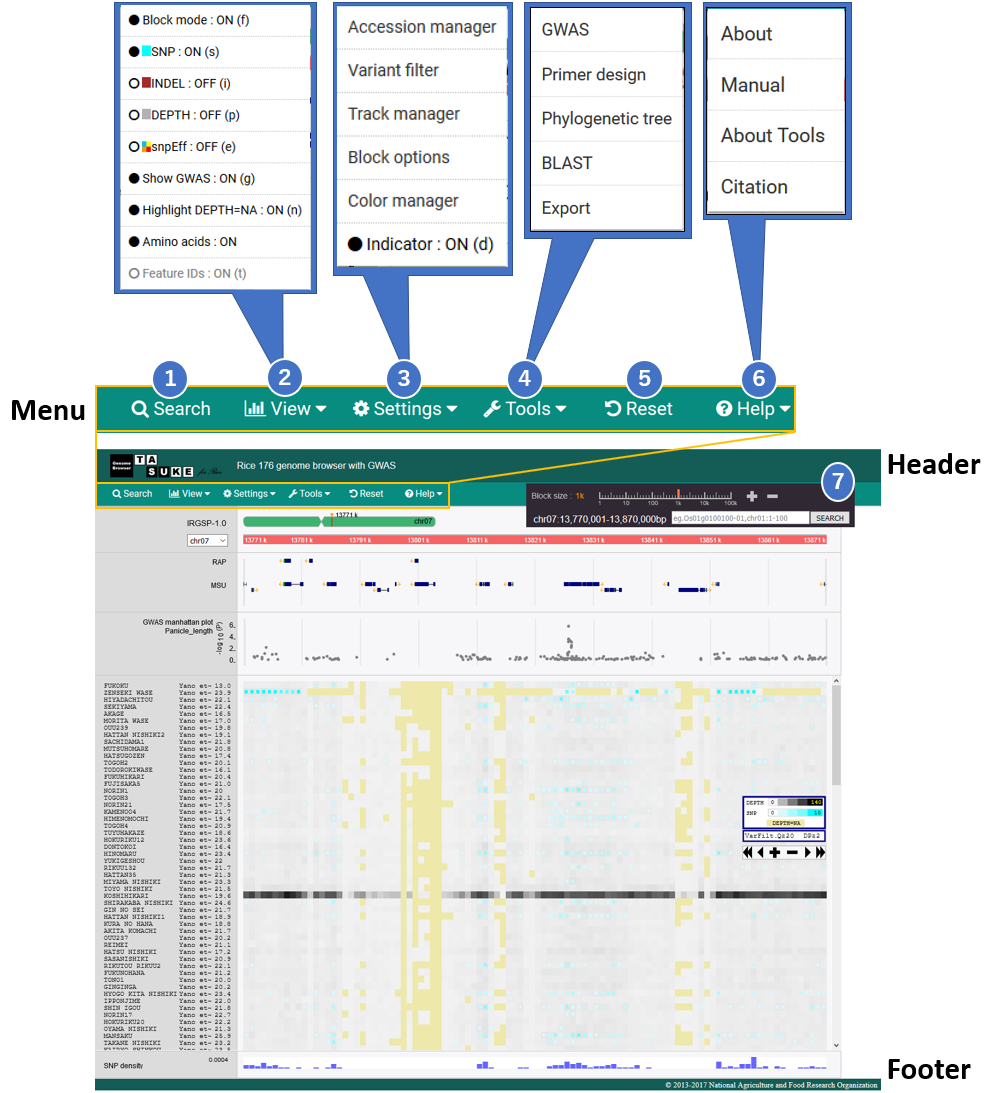
(a) Menu bar for various functions.
(b) Chromosomal positions.
(c) Annotation tracks.
(d) GWAS manhattan plot.
(e) Sample names and related information.
(f) Main panel for variant frequencies and average read depths of block regions.
(g) Magnified view of blocks. Blocks without reads are yellow
(h) Indicator for variant frequency and depth.
(i) Overall SNP density.
(j) Zoom bar and Search bar.
(k) Variants and average read depth information are shown by clicking on a block.
(l) Variant effects are shown by clicking on the sub-window of (k).
(m) Amino acids and nucleotides on a reference genome.
(n) Variants and their effects.
(o) Indicator of levels of variant effects.
Main Functions
Search the position by physical position, annotation ID or free words.
TASUKE shows Variant and Depth information with various scales, 1bp to 100kb per block. This level can be changed in Block size menu or simply double click on the position to be zoomed in the main field. Short cut keys ‘+’ and ‘↑’ for zoom in, and ‘‐‘ and ‘↓’ for zoom out are available.
View mode can be selected from "Frequency(Block)" or "Absolute position(In pixels)". In Absolute position mode, you can select between allele or GT display. "Depth", "SNP", and "INDEL" button switch the view of SNP block. You can custermize the SNP block on your purpose.
Variant Effect information added by snpEff can be shown by clicking the "View"-"snpEff" menu. "About" menu under "snpEff" menu refers the correspondence of colors and levels of effects.The indicator in the main field also shows the relations.
GWAS dialog shows the variant informa tion of each position. When you click the Link in "Position" , the GWAS plot on main view move this position.
Several genomic analysis from "Tools" menu.
Primer pairs can be designed by using Primer3. "Phylogenetic tree" get pylogenetic tree used PHYLIP. "BLAST" can do BLAST search used ncbi blast+ tools.
An image of currently shown region can be exported as <.png> file from “Export” menu. And FASTA and a variant list of selected region can be downloaded. If you are viewing a System phylogenetic tree, you can export the current tree in Newick format.
Basic actions and short cut keys are shown in ”Manual” window of “Help” menu.
Other options
Settings Menu-3
| Block options | Number of blocks, its height and width can be adjusted. |
|---|---|
| Accession/Tree manager | Accessions and optional information such as origin and variety name can be changed. |
| Track manager | Tracks to be shown, their order, color of the structure and number of rows can be changed. |
| Color manager | Block color definition and color gradient can be changed. |
| Indicator ON/OFF | Show and hide the indicator of depth and variant frequencies. |
| Cursor lines ON/OFF | Show and hide the cursor lines that follows the mouse on the track. This makes it easier to compare the positions of variants and genes. |
| Variant Filter Menu-3 | Filter variants by depth of all reads, alternative (non-reference type) allele reads and genotyping quality. |
|---|---|
| Highlight DEPTH=NULL ON/OFF Menu-2 | Highlight the depth = null (unmapped) site. |
Reset all the setting to default values.