How to use (if using the previous version, check here)
Views
Tracks


Frequency Mode
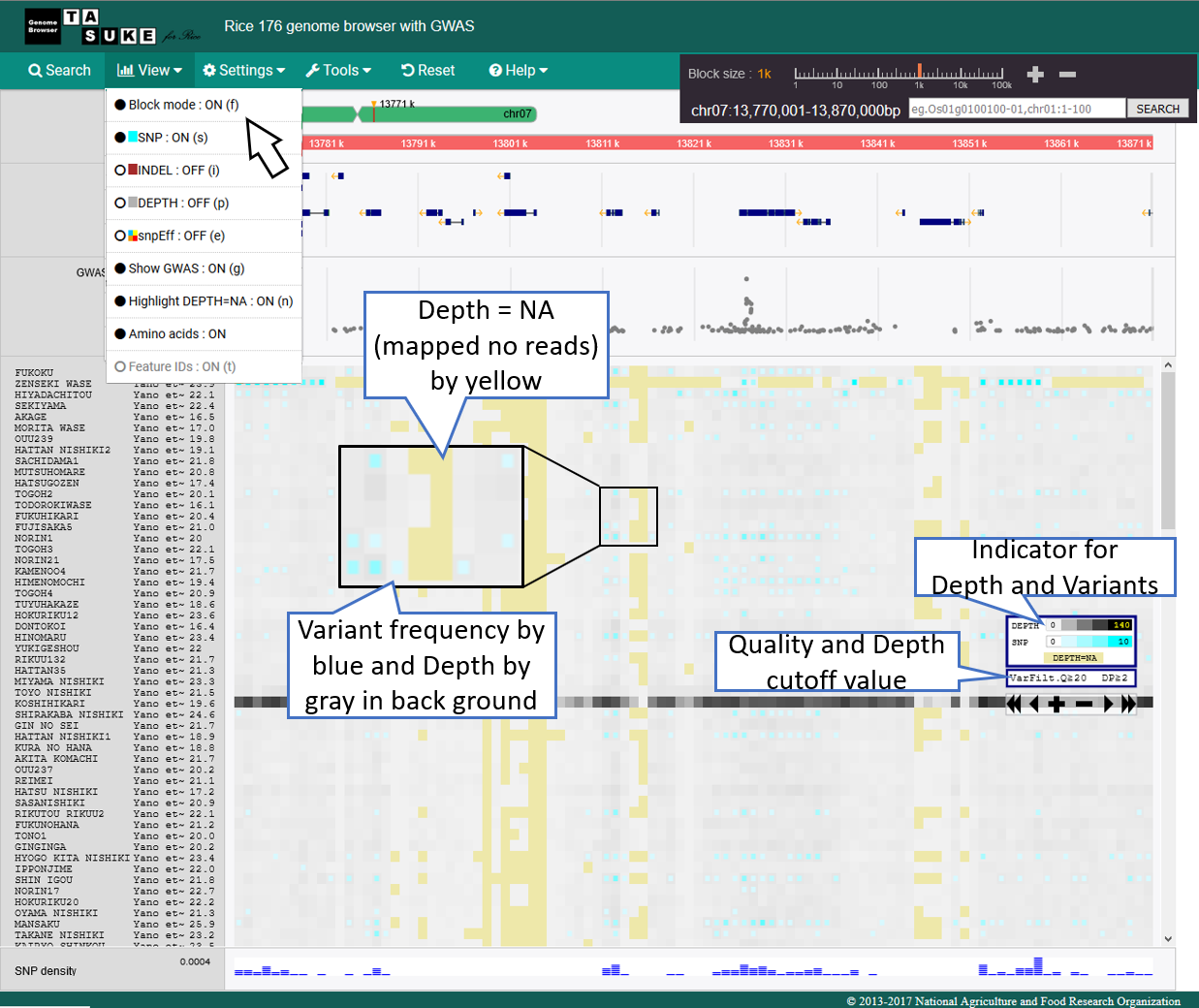

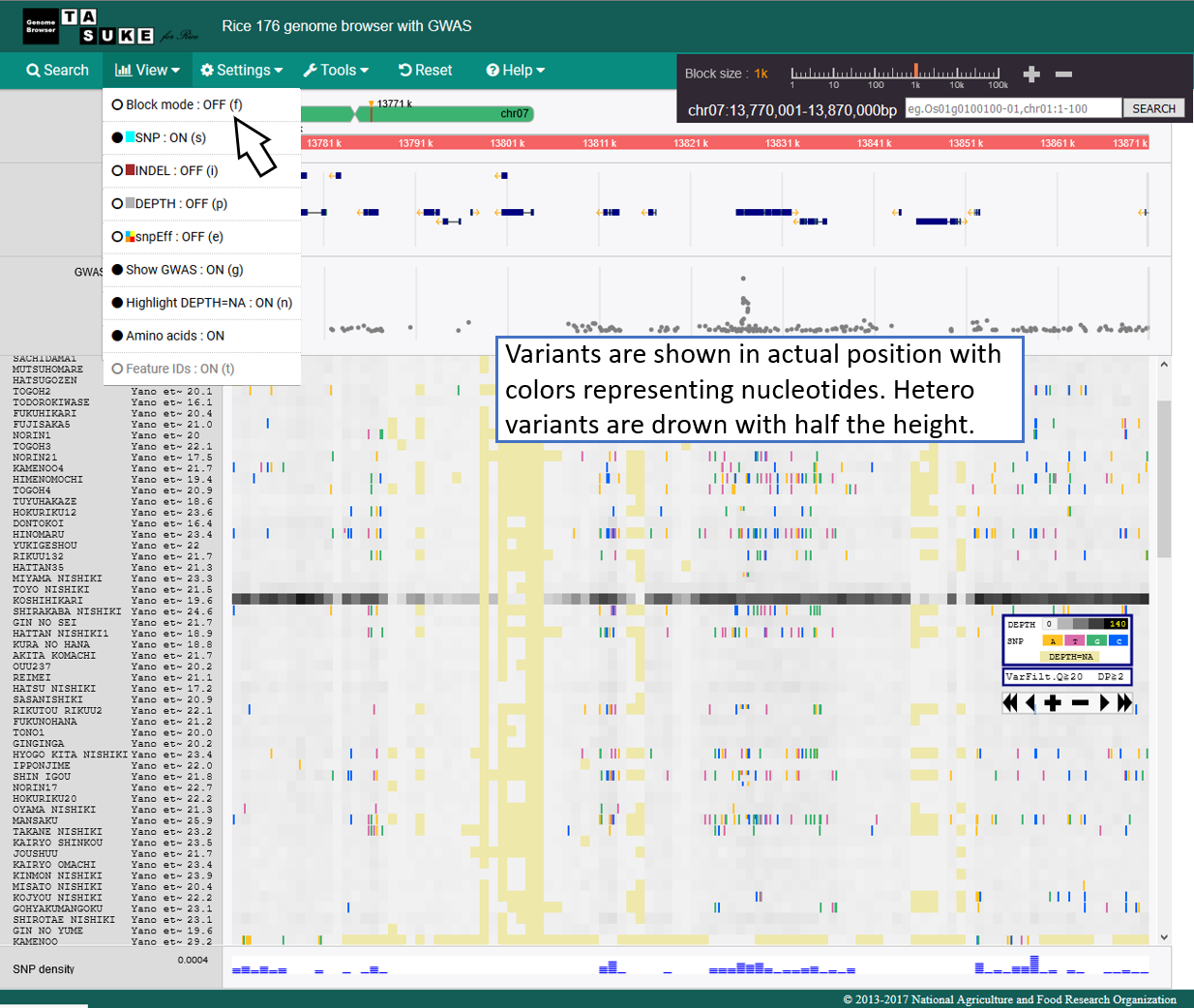
Absolute position mode
snpEff Mode
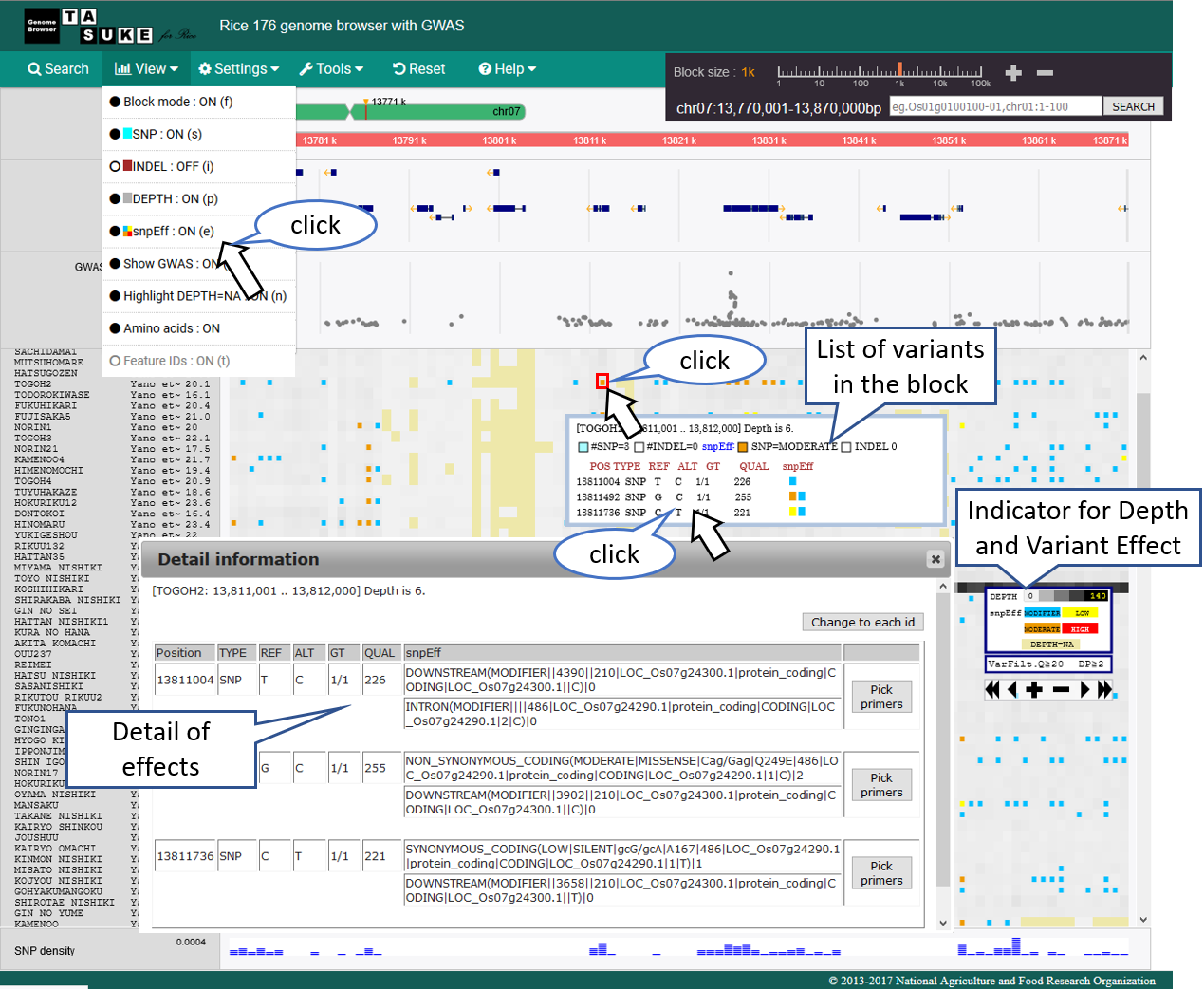
Effect category

View of 1bp Scale

Annotation track
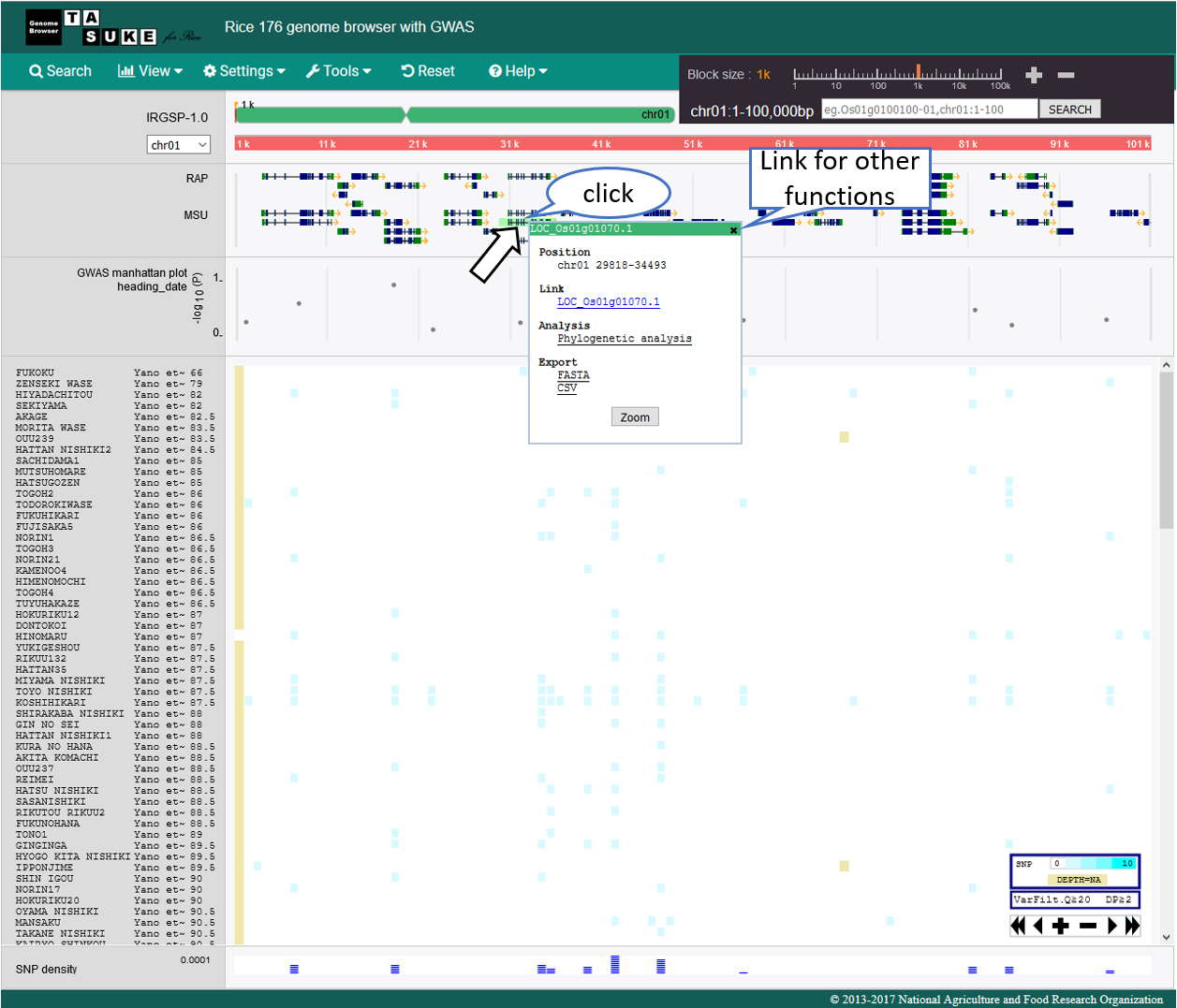
Action
Search & Jump
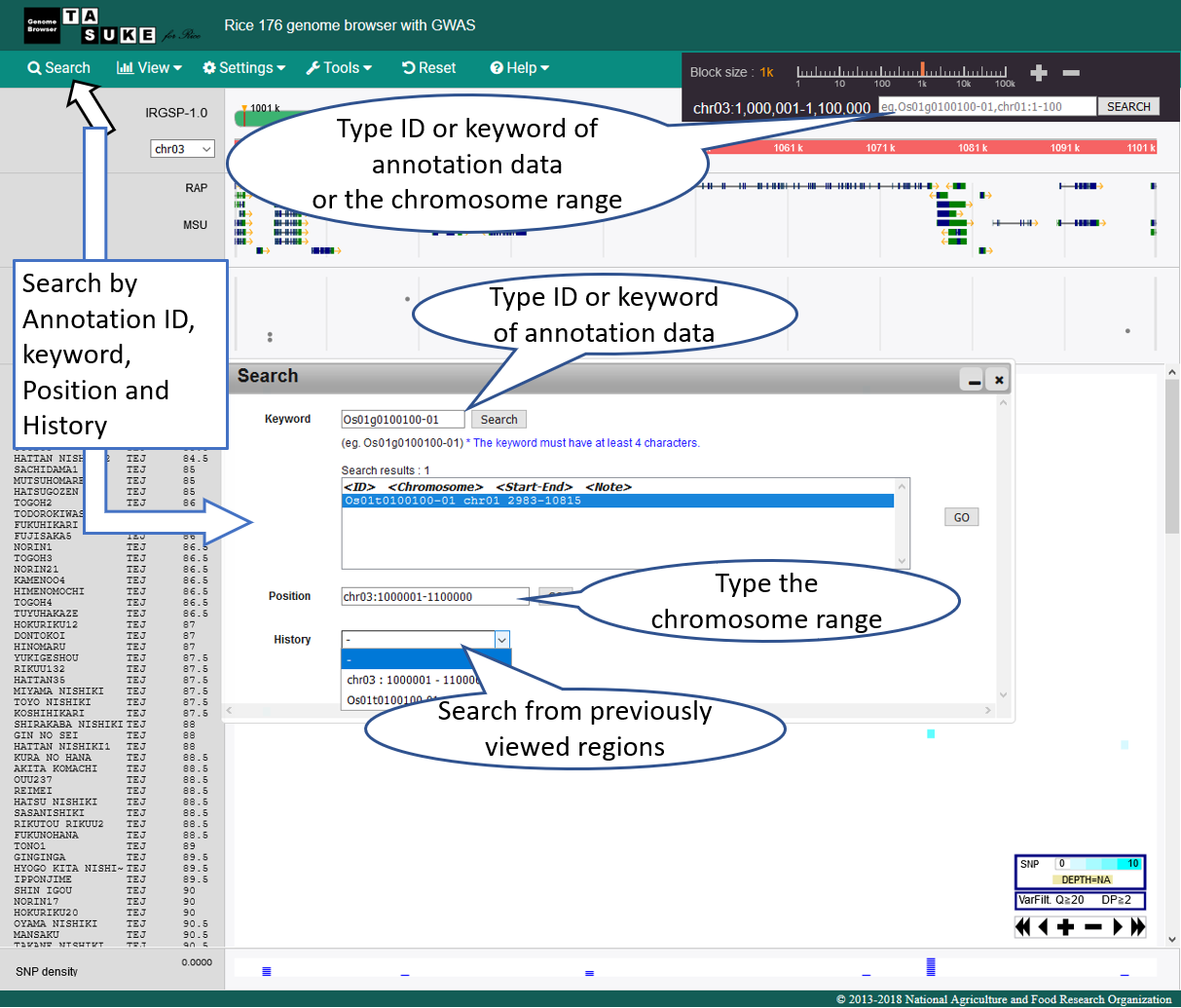

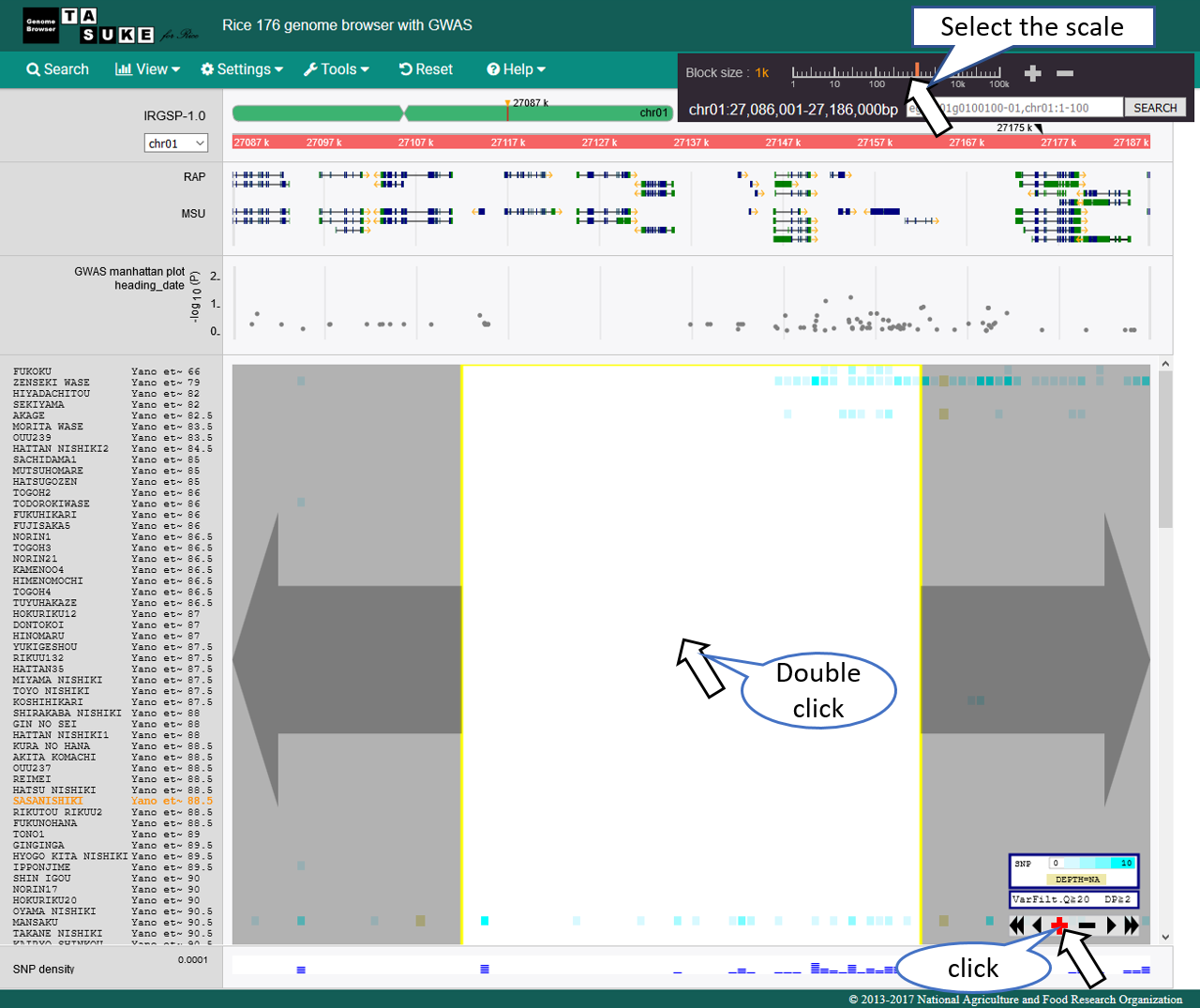
Move


Zoom

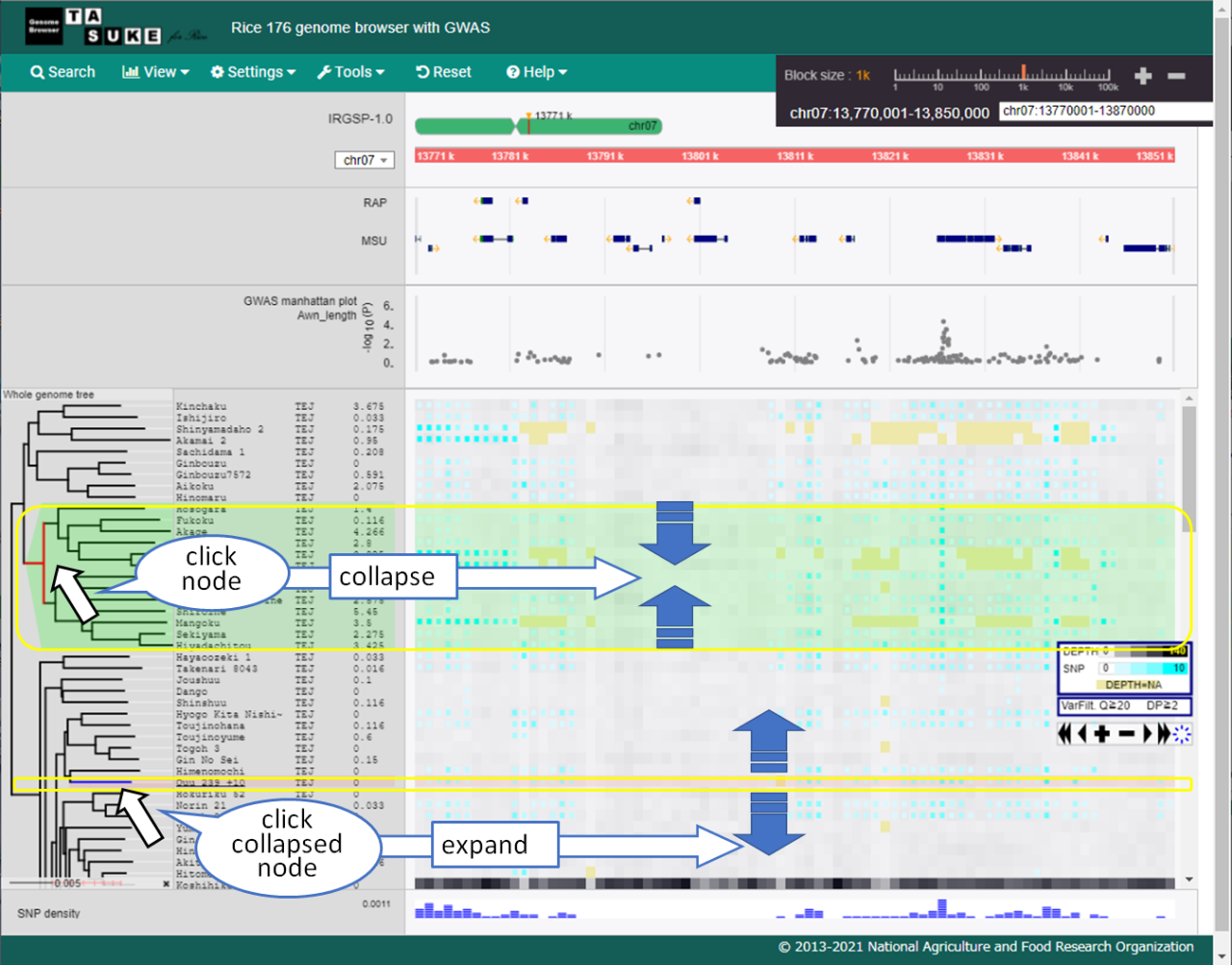
System phylogenetic tree

Tools
Design PRIMER
1.Open setting dialog

2.Parameter setting
3.Result
1.Open setting dialog

2.Parameter setting
2-1.Main settings
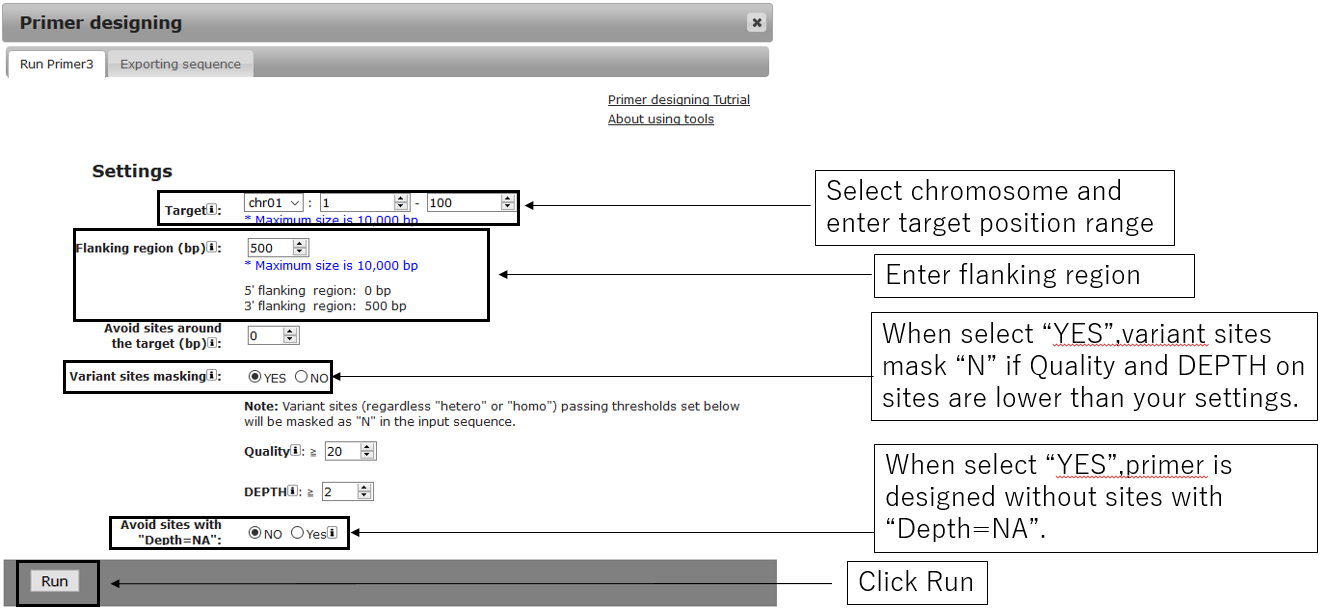
2-2.General settings(Optional)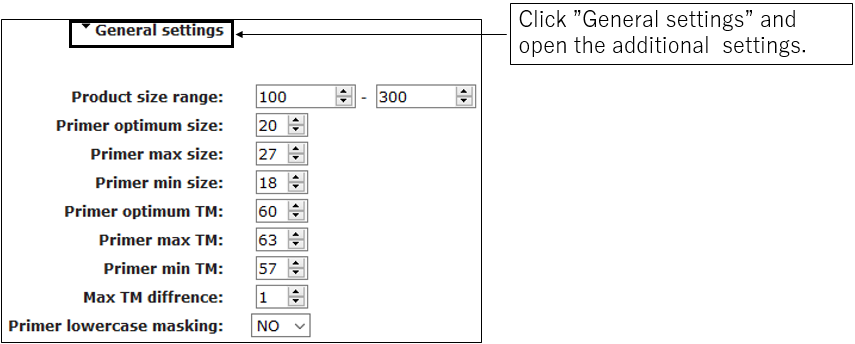
2-3.ePCR settings(Optional)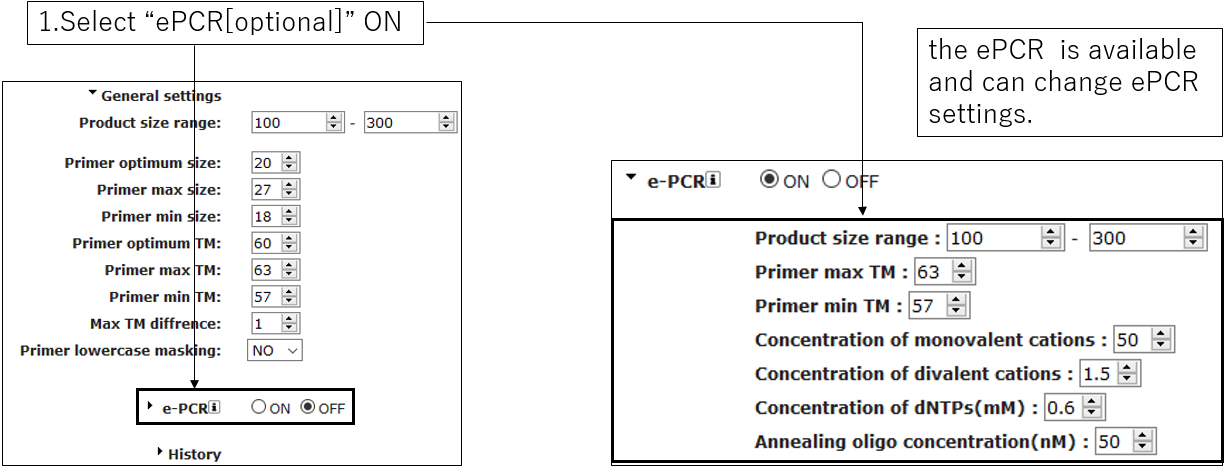

2-2.General settings(Optional)

2-3.ePCR settings(Optional)

3.Result
3-1.Result

3-2.Jump BLAST with primer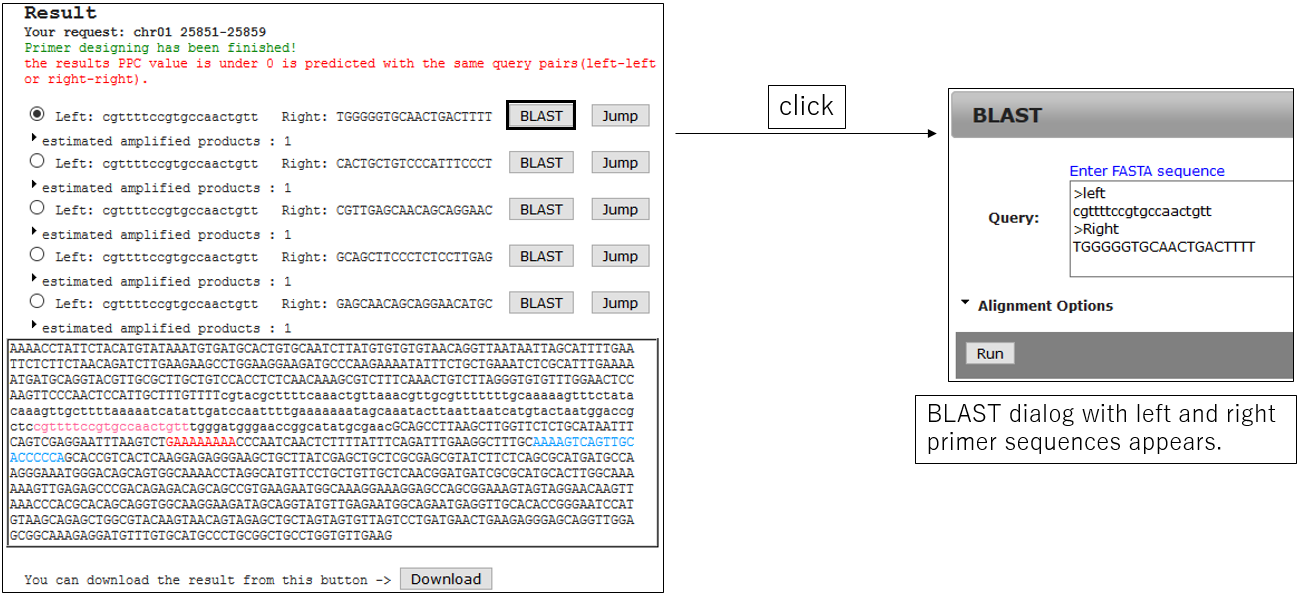
3-3.Move to the perimer designing position
3-4.ePCR Result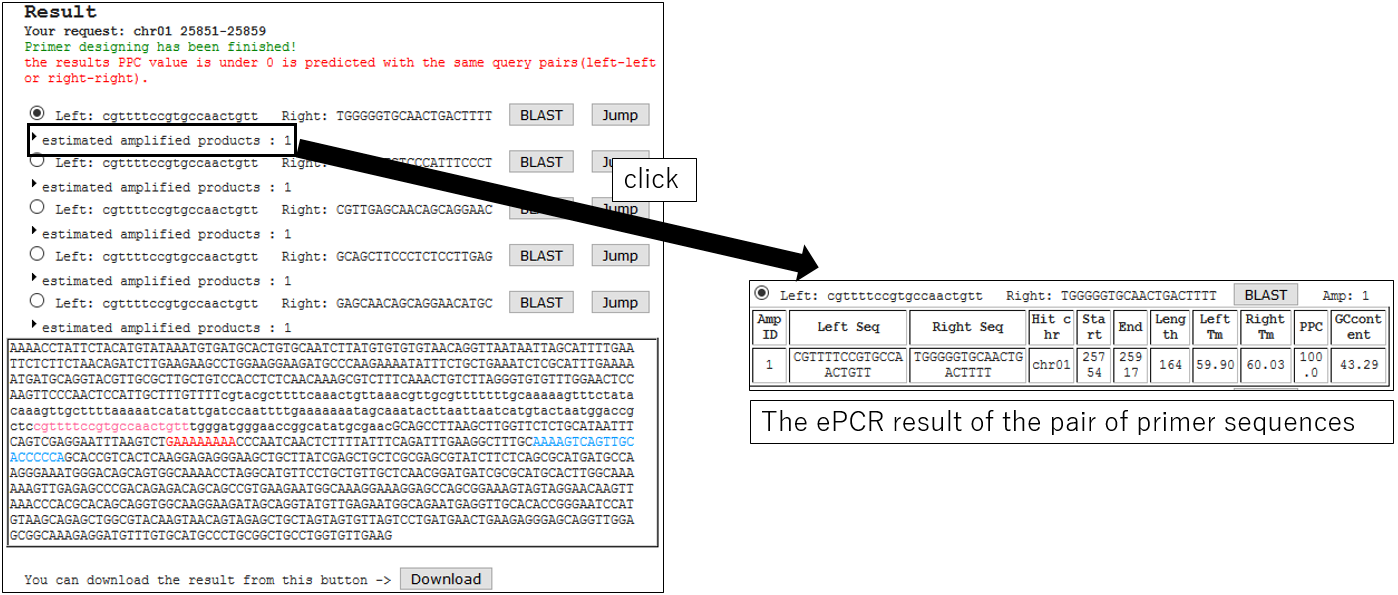
3-5.Download Result
3-6.Download Result(using ePCR)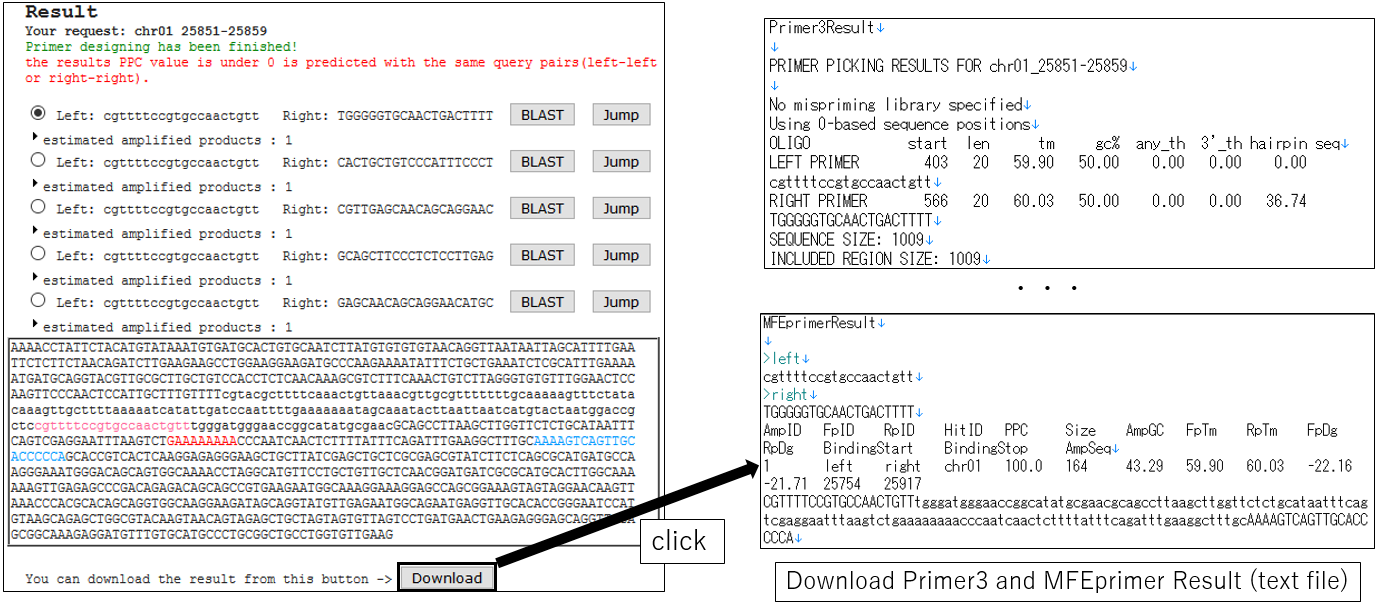

3-2.Jump BLAST with primer

3-3.Move to the perimer designing position

3-4.ePCR Result

3-5.Download Result

3-6.Download Result(using ePCR)

Create Phylogenetic Tree
If the accession name is set to "Name" display, the phylogenetic tree may not be displayed correctly. If so, change it to "ID" in the AccessionManager.
1.Open setting dialog
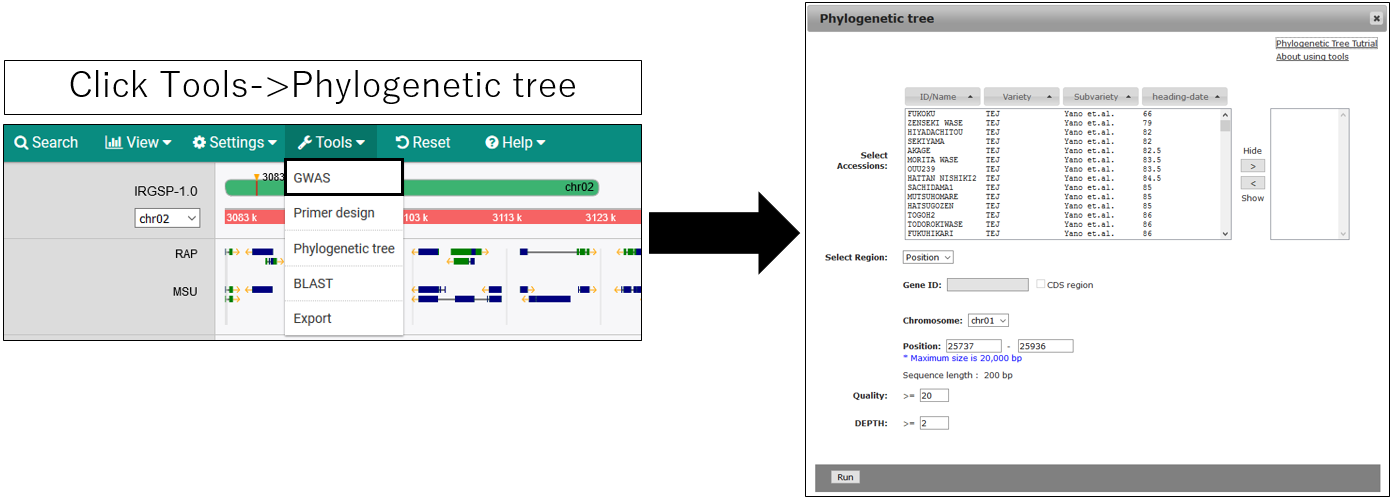
2.Parameter setting
If the accession name is set to "Name" display, the phylogenetic tree may not be displayed correctly. If so, change it to "ID" in the AccessionManager.
1.Open setting dialog

2.Parameter setting
2-1.Main settings(with Chromosome and Position)
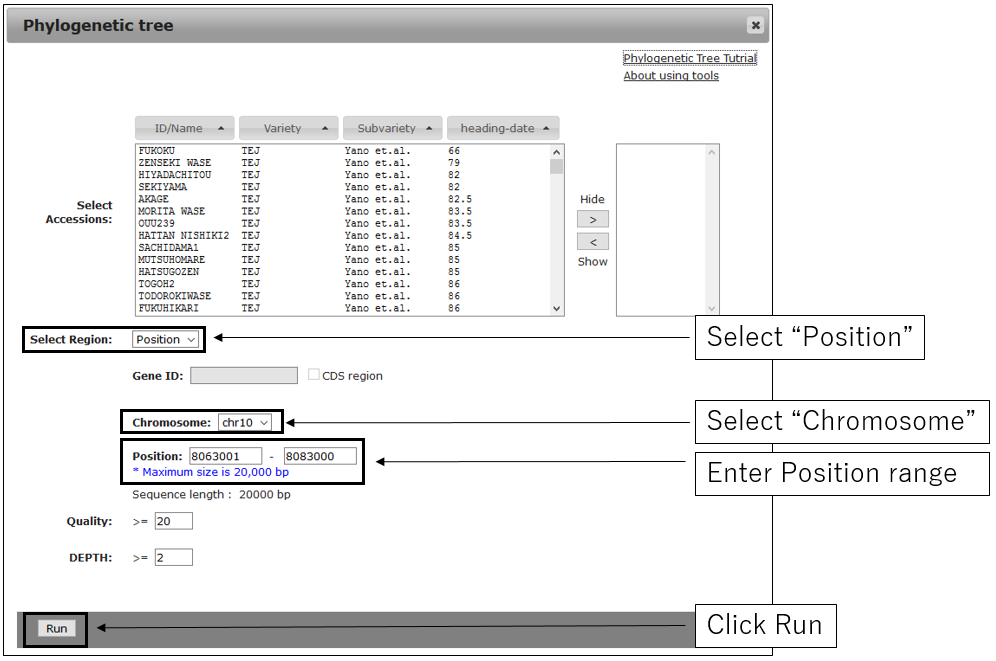
2-2.Main settings(with GeneID)
3.Result

2-2.Main settings(with GeneID)

3-1.Result
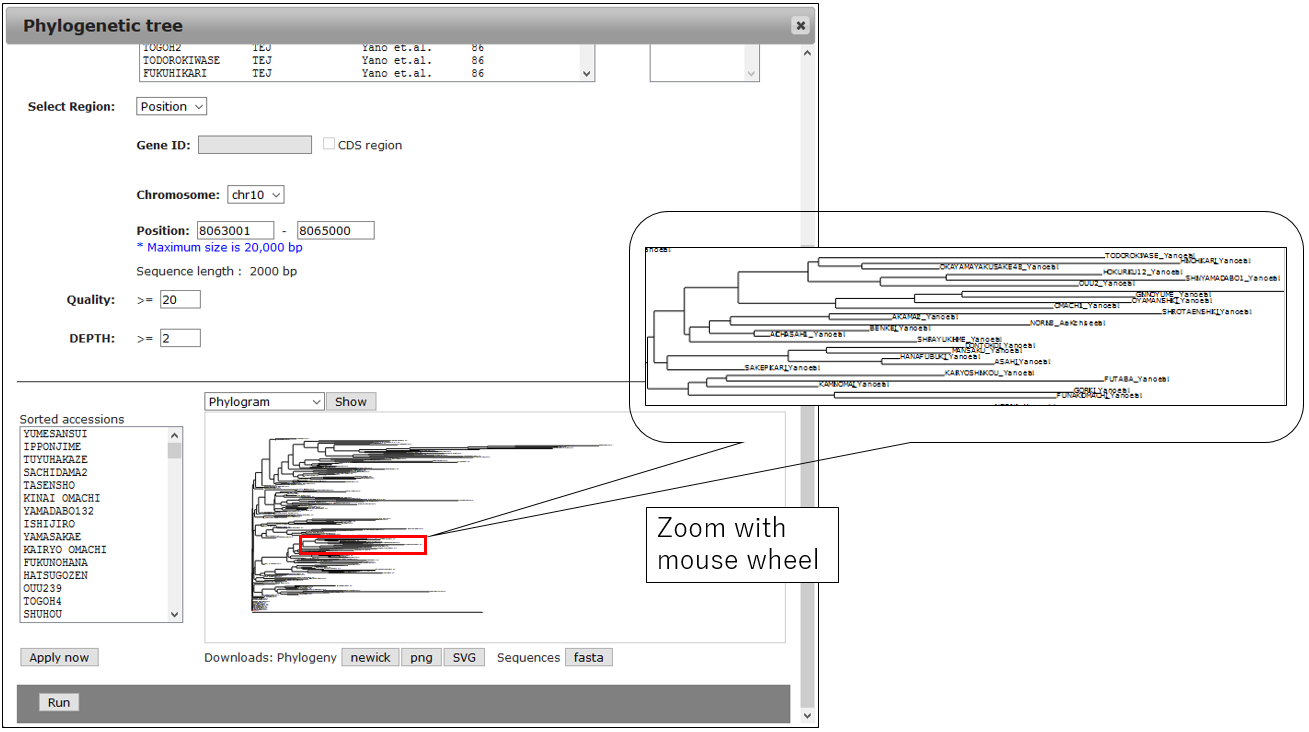
3-2.Change Circle phylogram
3-3.Change Cladogram
3-4.Sort snpblock table
3-5.Download tree data
3-6.Select Accession (option)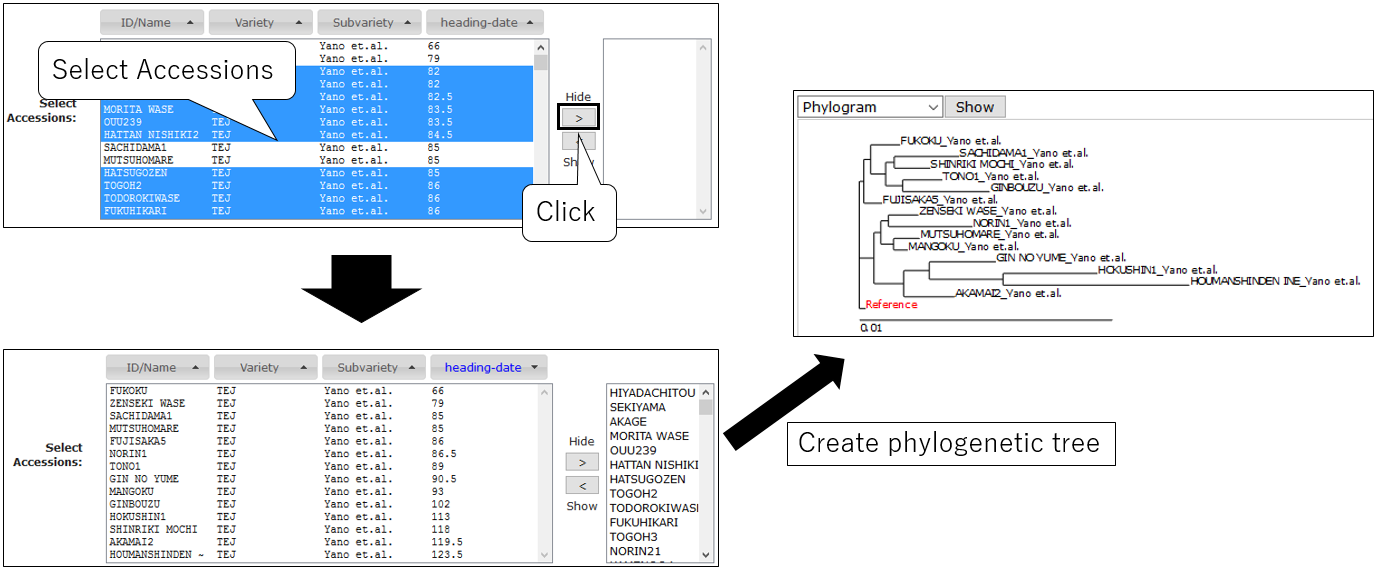

3-2.Change Circle phylogram

3-3.Change Cladogram

3-4.Sort snpblock table

3-5.Download tree data

3-6.Select Accession (option)

BLAST
1.Open setting dialog
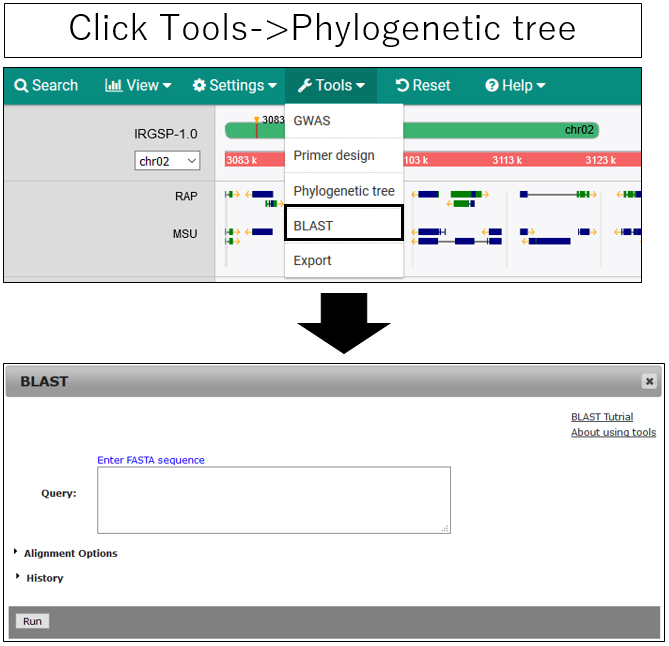
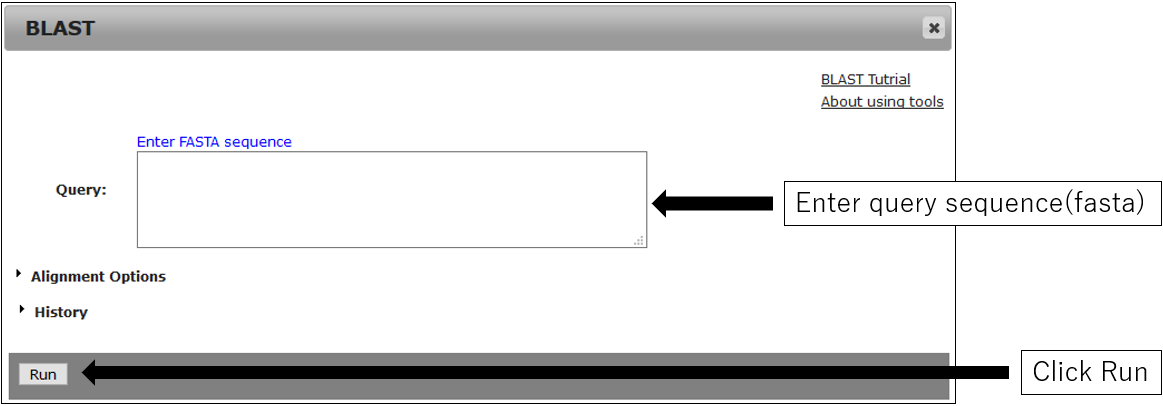
2.Parameter setting
1.Open setting dialog

2.Parameter setting
2-1.Main settings
2-2.Alignment Options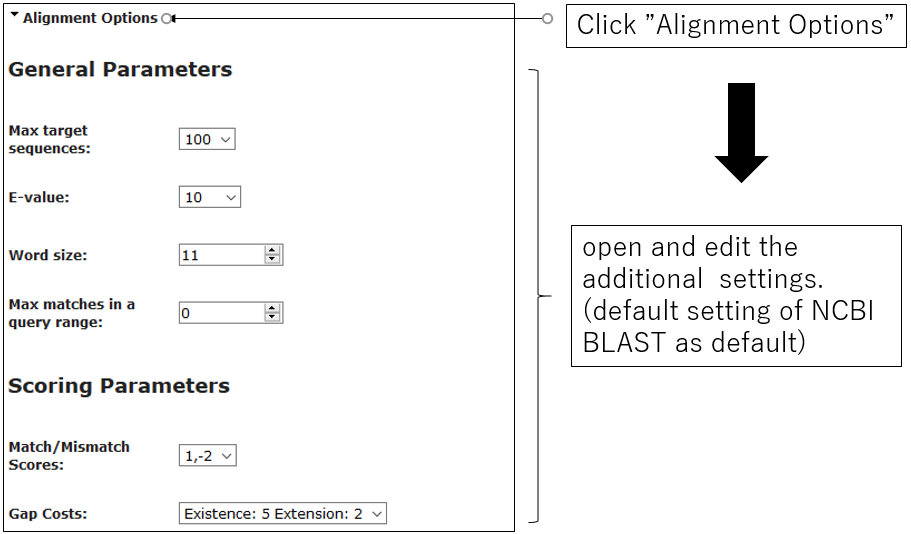
3.Result

2-2.Alignment Options

3-1.Result
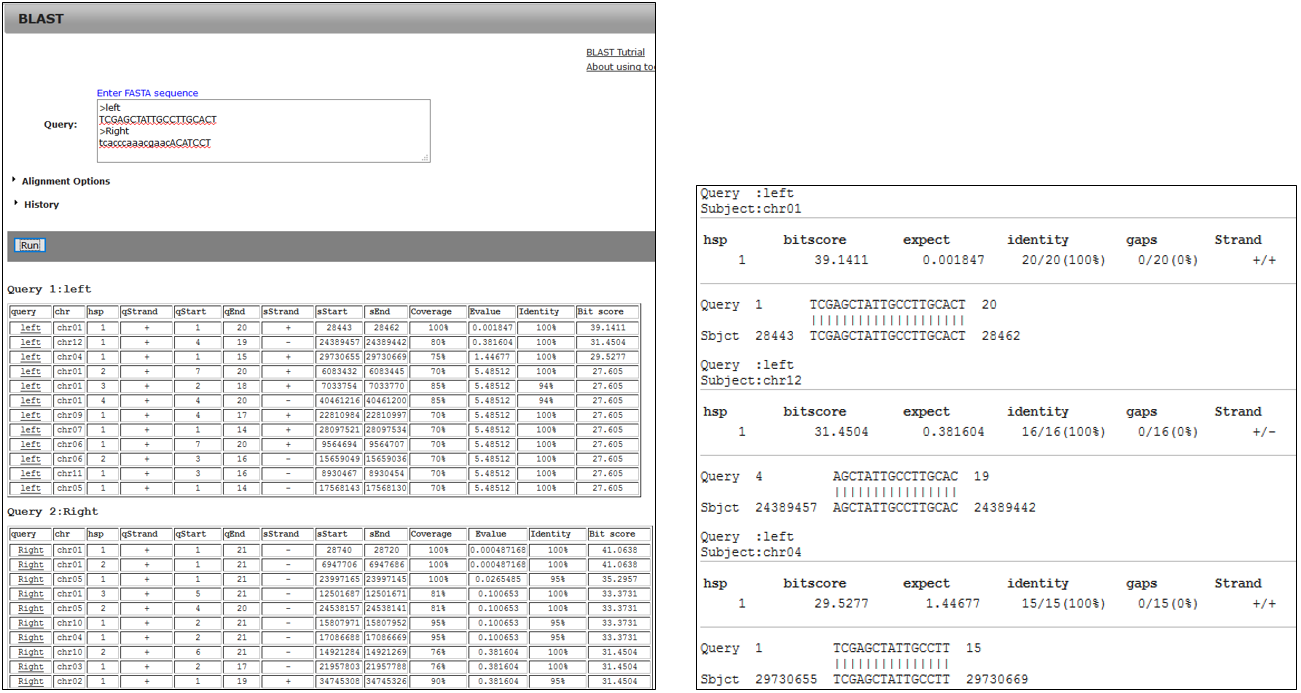
3-2.BLAST result using primer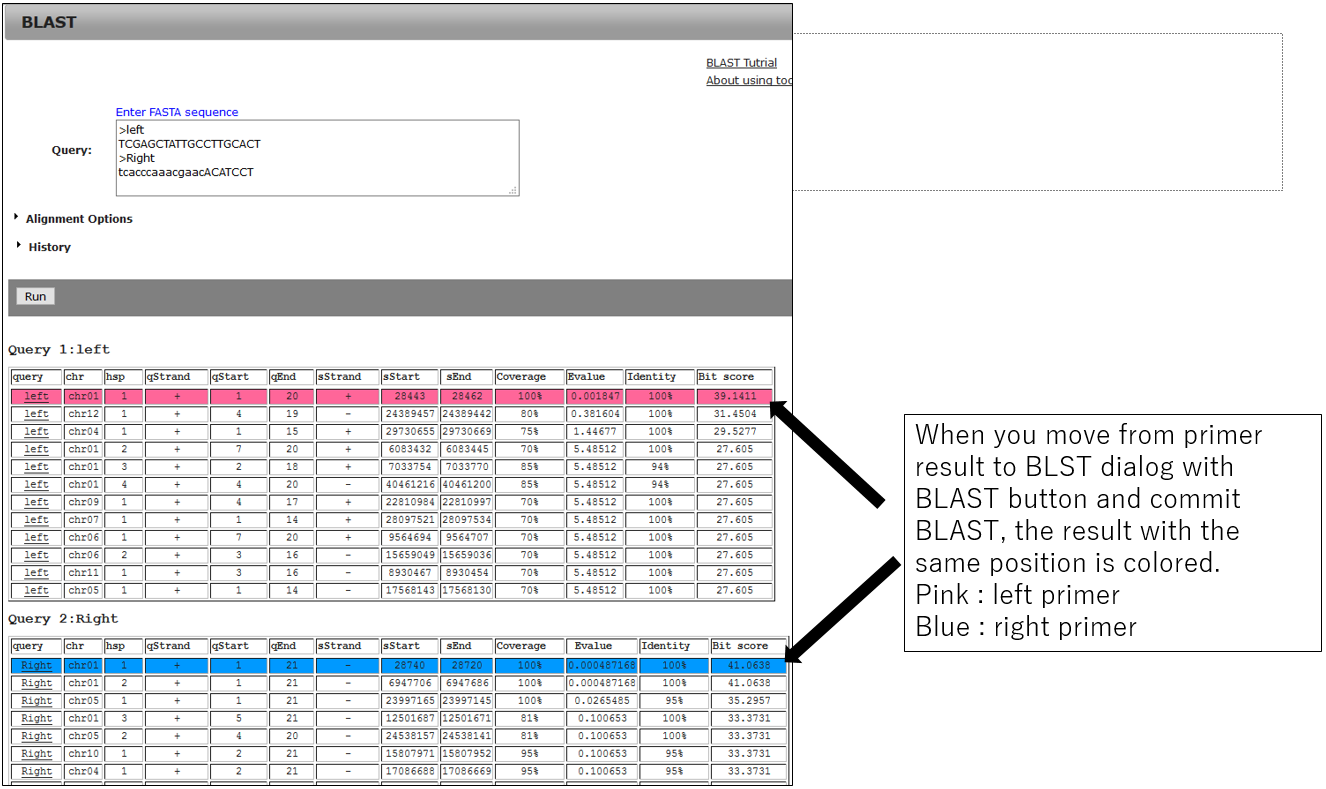
3-3.Jump alignment section of BLAST result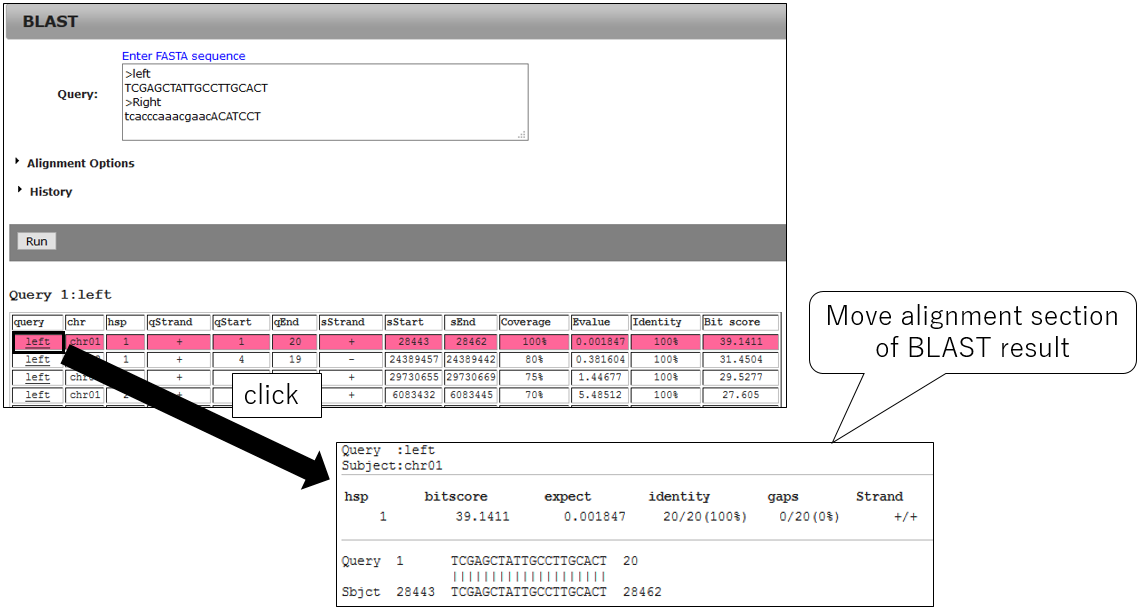

3-2.BLAST result using primer

3-3.Jump alignment section of BLAST result

GWAS
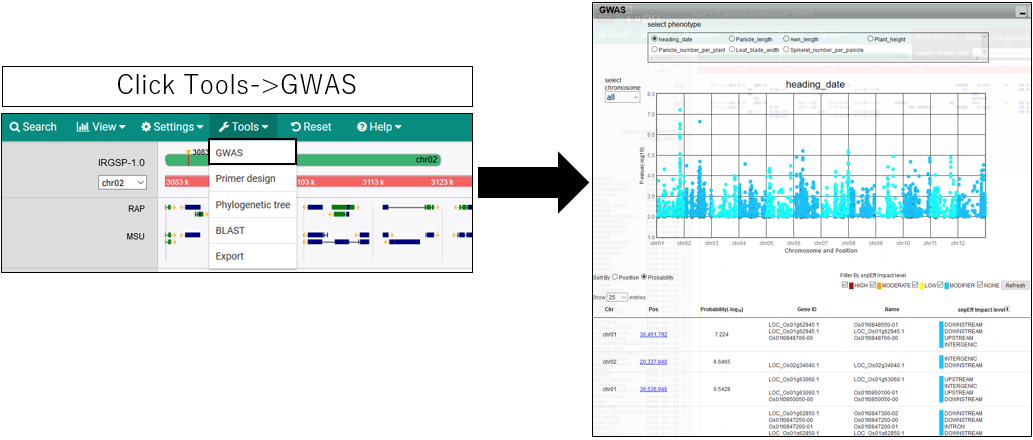
1.Open GWAS dialog
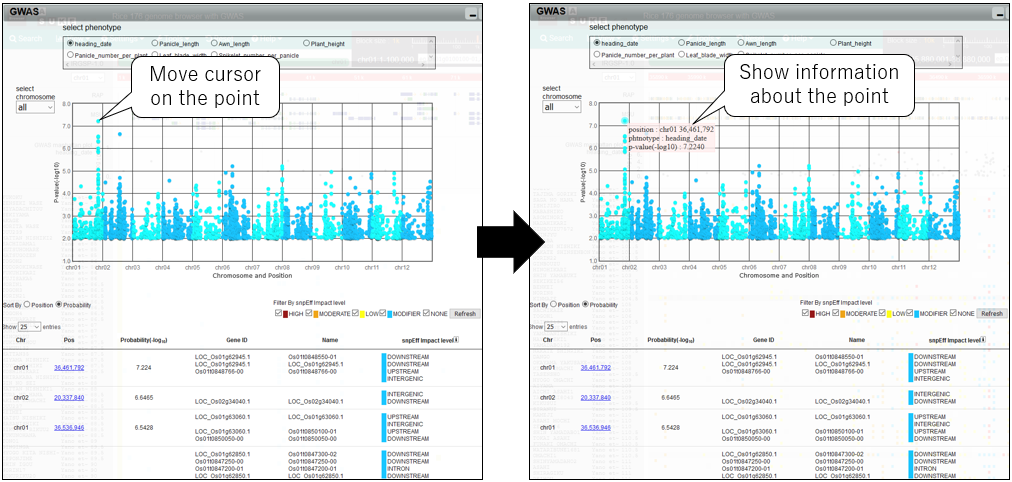
2.Show information
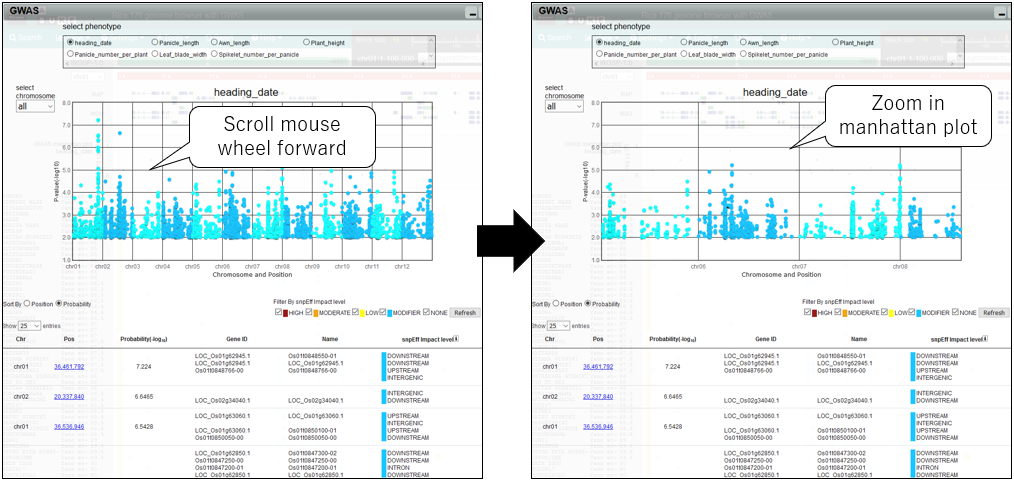
3.Zoom in manhattan plot
4.Zoom out manhattan plot

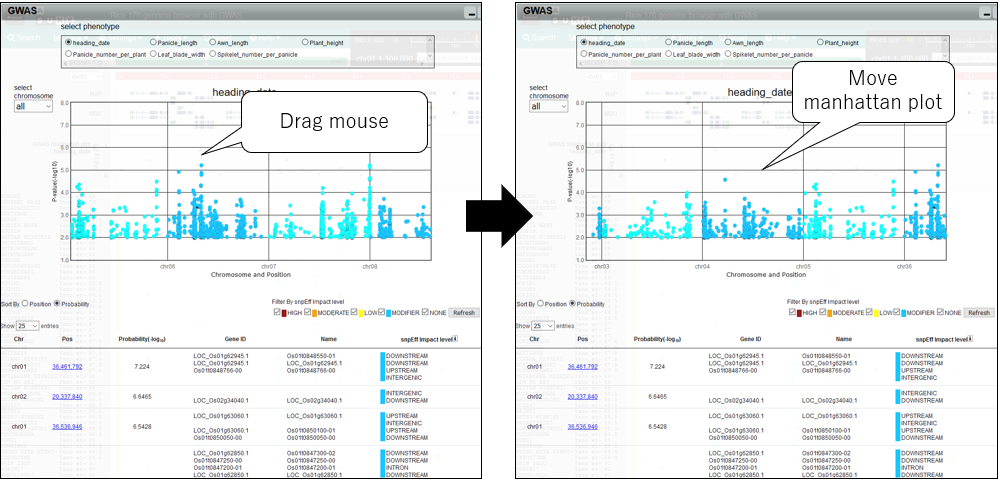
5.Pan manhattan plot
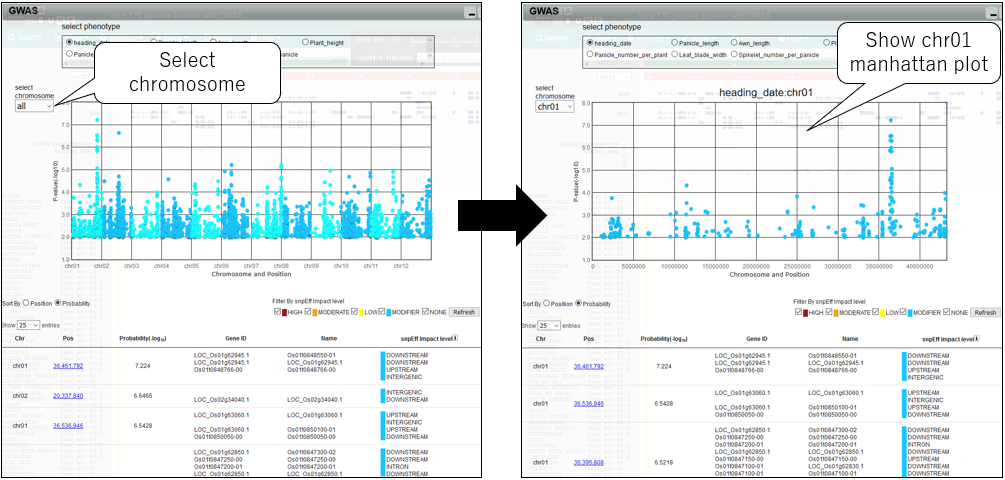
6.Change chromosome
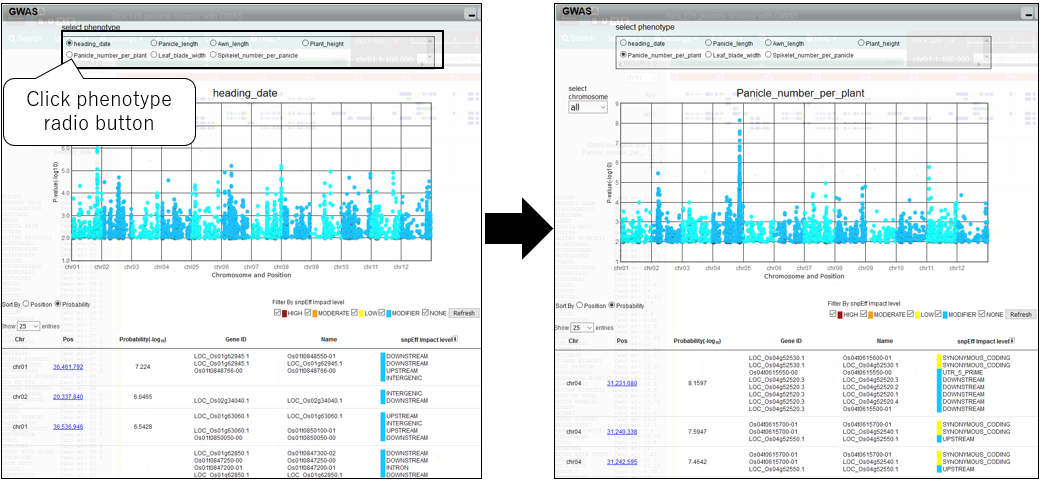
7.Change phenotype
8.Filter By SnpEff impact

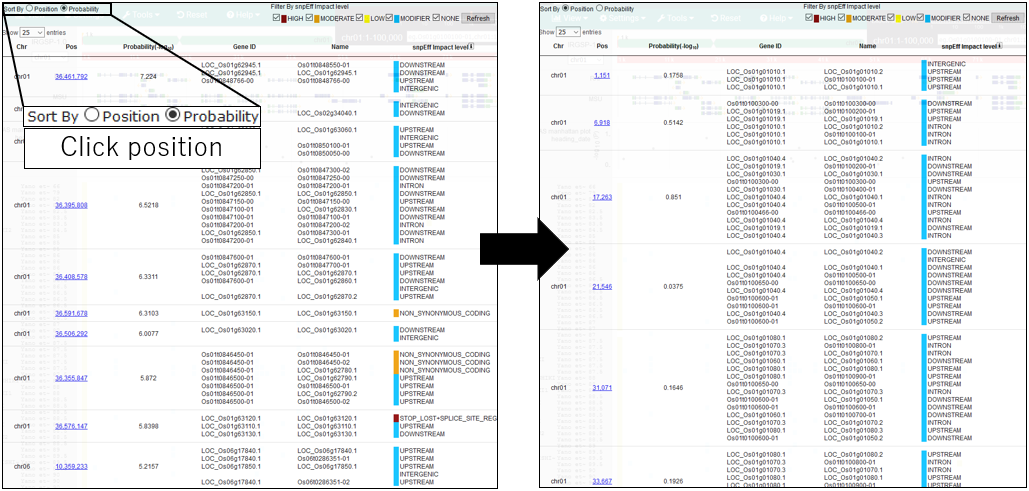
9.Sort By Position
10.Move postiion on GWAS plot

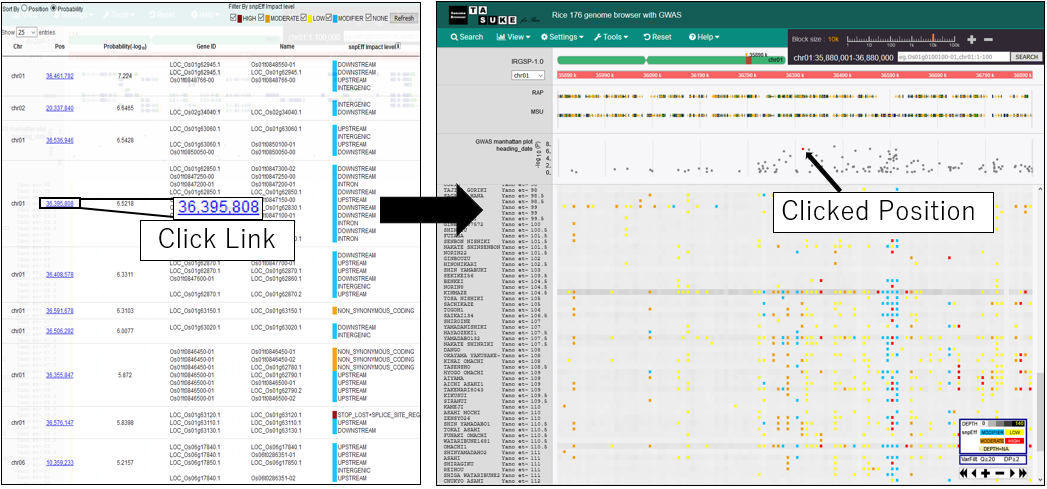
11.Move postiion on variant list
1.Open GWAS dialog

2.Show information

3.Zoom in manhattan plot

4.Zoom out manhattan plot

5.Pan manhattan plot

6.Change chromosome

7.Change phenotype

8.Filter By SnpEff impact

9.Sort By Position

10.Move postiion on GWAS plot

11.Move postiion on variant list

